表の作成は大変時間がかかり手間なものです。そんな問題を解決するパッケージの紹介です。本パッケージには基本の表を作成する「tbl_summary」コマンド、回帰モデルの結果を表にする「tbl_regression」コマンド、「表の体裁」や「各種検定を実施し表に追加」するコマンドが多数収録されています。なお、表のファイルへの出力はRStudioを利用すると簡単です。ファイルは画像やhtmlで保存が可能です。
紹介では基本の表を作成する「tbl_summary」コマンド 例とパッケージで 実施可能な検定をまとめました。
パッケージバージョンは1.5.1。実行コマンドはwindows 11のR version 4.1.2で確認しています。
パッケージのインストール
下記コマンドを実行してください。
#パッケージのインストール
install.packages("gtsummary")実行コマンド
詳細はコマンド、パッケージのヘルプを確認してください。
#パッケージの読み込み
library("gtsummary")
#tidyverseパッケージがなければインストール
if(!require("tidyverse", quietly = TRUE)){
install.packages("tidyverse");require("tidyverse")
}
###データ例の作成#####
set.seed(1234)
TestData <- tibble(Group = sample(paste0("Group", 1:2), 100,
replace = TRUE),
Data1 = sample(50:200, 100, replace = TRUE),
Data2 = sample(0:10, 100, replace = TRUE),
Data3 = sample(LETTERS[1:3], 100, replace = TRUE))
#確認
TestData
# A tibble: 100 x 4
Group Data1 Data2 Data3
<chr> <int> <int> <chr>
1 Group2 145 8 A
2 Group1 132 6 C
3 Group2 179 4 B
4 Group2 56 8 C
5 Group1 136 5 B
6 Group1 149 1 A
7 Group2 174 0 A
8 Group1 55 0 A
9 Group1 64 6 C
10 Group2 188 4 C
# ... with 90 more rows
########
#データをテーブルに変換:tbl_summaryコマンド
#単純に変換
TestData %>%
tbl_summary()
#Groupで分割し変換
#分割指標を設定:byオプション
TestData %>%
tbl_summary(by = Group) %>%
#列を追加
add_n() %>%
#検定結果を追加:add_pオプション
#各種検定の検定コマンドに引数を渡す:test.argsオプション
#例では分散が等しいと仮定して"t.test"を求める
#各種検定詳細は記事内を確認くしてください
add_p(test = list(all_continuous() ~ "t.test",
Data3 ~ "chisq.test"),
test.args = all_continuous() ~ list(var.equal = TRUE)) %>%
#先頭行の内容設定:modify_headerオプション
#**で囲むと太字になります,以下同様
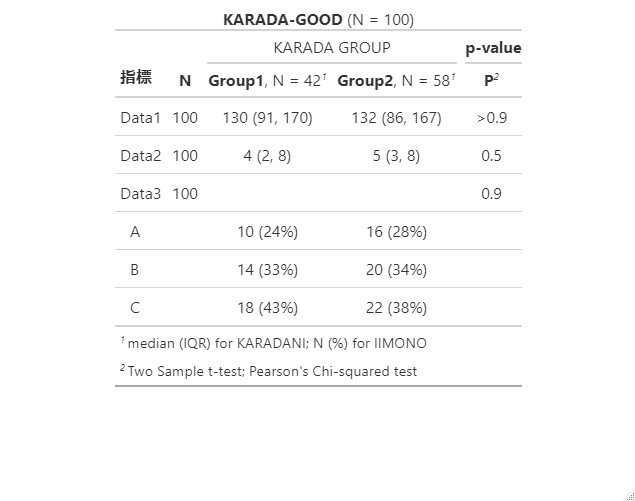
modify_header(label = "**指標**", p.value = "**P**") %>%
#先頭行を分割する
modify_spanning_header(list(all_stat_cols() ~ "KARADA GROUP",
starts_with("p.value") ~ "**p-value**")) %>%
#脚注の内容設定:modify_footnoteオプション
modify_footnote(all_stat_cols() ~ "median (IQR) for KARADANI; N (%) for IIMONO") %>%
#表題の内容設定:modify_footnoteオプション
modify_caption("**KARADA-GOOD** (N = {N})")出力例
実施可能な各種検定
左から適応可能なコマンド、add_p(test)に設定する書式、説明、バックで動作している検定コマンドです。パッケージヘルプを参照し作成しました。
| 適応可能コマンド | X | 説明 | 検定コマンド |
|---|---|---|---|
| tbl_summary() %>% add_p(test = X) | "t.test" | t-test | t.test(variable ~ as.factor(by), data = data, conf.level = 0.95, ...) |
| tbl_summary() %>% add_p(test = X) | "aov" | One-way ANOVA | aov(variable ~ as.factor(by), data = data) %>% summary() |
| tbl_summary() %>% add_p(test = X) | "kruskal.test" | Kruskal-Wallis test | kruskal.test(data[[variable]], as.factor(data[[by]])) |
| tbl_summary() %>% add_p(test = X) | "wilcox.test" | Wilcoxon rank-sum test | wilcox.test(as.numeric(variable) ~ as.factor(by), data = data, ...) |
| tbl_summary() %>% add_p(test = X) | "chisq.test" | chi-square test of independence | chisq.test(x = data[[variable]], y = as.factor(data[[by]]), ...) |
| tbl_summary() %>% add_p(test = X) | "chisq.test.no.correct" | chi-square test of independence | chisq.test(x = data[[variable]], y = as.factor(data[[by]]), correct = FALSE) |
| tbl_summary() %>% add_p(test = X) | "fisher.test" | Fisher's exact test | fisher.test(data[[variable]], as.factor(data[[by]]), conf.level = 0.95, ...) |
| tbl_summary() %>% add_p(test = X) | "mcnemar.test" | McNemar's test | tidyr::pivot_wider(id_cols = group, ...); mcnemar.test(by_1, by_2, conf.level = 0.95, ...) |
| tbl_summary() %>% add_p(test = X) | "mcnemar.test.wide" | McNemar's test | mcnemar.test(data[[variable]], data[[by]], conf.level = 0.95, ...) |
| tbl_summary() %>% add_p(test = X) | "lme4" | random intercept logistic regression | lme4::glmer(by ~ (1 \UFF5C group), data, family = binomial) %>% anova(lme4::glmer(by ~ variable + (1 \UFF5C group), data, family = binomial)) |
| tbl_summary() %>% add_p(test = X) | "paired.t.test" | Paired t-test | tidyr::pivot_wider(id_cols = group, ...); t.test(by_1, by_2, paired = TRUE, conf.level = 0.95, ...) |
| tbl_summary() %>% add_p(test = X) | "paired.wilcox.test" | Wilcoxon rank-sum test | tidyr::pivot_wider(id_cols = group, ...); wilcox.test(by_1, by_2, paired = TRUE, conf.int = TRUE, conf.level = 0.95, ...) |
| tbl_summary() %>% add_p(test = X) | "prop.test" | Test for equality of proportions | prop.test(x, n, conf.level = 0.95, ...) |
| tbl_summary() %>% add_p(test = X) | "ancova" | ANCOVA | lm(variable ~ by + adj.vars) |
| tbl_summary() %>% add_p(test = X) | "emmeans" | Estimated Marginal Means or LS-means | lm(variable ~ by + adj.vars, data) %>% emmeans::emmeans(specs =~by) %>% emmeans::contrast(method = "pairwise") %>% summary(infer = TRUE, level = conf.level) |
| tbl_svysummary() %>%add_p(test = X) | "svy.t.test" | t-test adapted to complex survey samples | survey::svyttest(~variable + by, data) |
| tbl_svysummary() %>%add_p(test = X) | "svy.wilcox.test" | Wilcoxon rank-sum test for complex survey samples | survey::svyranktest(~variable + by, data, test = 'wilcoxon') |
| tbl_svysummary() %>%add_p(test = X) | "svy.kruskal.test" | Kruskal-Wallis rank-sum test for complex survey samples | survey::svyranktest(~variable + by, data, test = 'KruskalWallis') |
| tbl_svysummary() %>%add_p(test = X) | "svy.vanderwaerden.test" | van der Waerden's normal-scores test for complex survey samples | survey::svyranktest(~variable + by, data, test = 'vanderWaerden') |
| tbl_svysummary() %>%add_p(test = X) | "svy.median.test" | Mood's test for the median for complex survey samples | survey::svyranktest(~variable + by, data, test = 'median') |
| tbl_svysummary() %>%add_p(test = X) | "svy.chisq.test" | chi-squared test with Rao & Scott's second-order correction | survey::svychisq(~variable + by, data, statistic = 'F') |
| tbl_svysummary() %>%add_p(test = X) | "svy.adj.chisq.test" | chi-squared test adjusted by a design effect estimate | survey::svychisq(~variable + by, data, statistic = 'Chisq') |
| tbl_svysummary() %>%add_p(test = X) | "svy.wald.test" | Wald test of independence for complex survey samples | survey::svychisq(~variable + by, data, statistic = 'Wald') |
| tbl_svysummary() %>%add_p(test = X) | "svy.adj.wald.test" | adjusted Wald test of independence for complex survey samples | survey::svychisq(~variable + by, data, statistic = 'adjWald') |
| tbl_svysummary() %>%add_p(test = X) | "svy.lincom.test" | test of independence using the exact asymptotic distribution for complex survey samples | survey::svychisq(~variable + by, data, statistic = 'lincom') |
| tbl_svysummary() %>%add_p(test = X) | "svy.saddlepoint.test" | test of independence using a saddlepoint approximation for complex survey samples | survey::svychisq(~variable + by, data, statistic = 'saddlepoint') |
| tbl_survfit() %>% add_p(test = X) | "logrank" | Log-rank test | survival::survdiff(Surv(.) ~ variable, data, rho = 0) |
| tbl_survfit() %>% add_p(test = X) | "petopeto_gehanwilcoxon" | Peto & Peto modification of Gehan-Wilcoxon test | survival::survdiff(Surv(.) ~ variable, data, rho = 1) |
| tbl_survfit() %>% add_p(test = X) | "survdiff" | G-rho family test | survival::survdiff(Surv(.) ~ variable, data, ...) |
| tbl_survfit() %>% add_p(test = X) | "coxph_lrt" | Cox regression (LRT) | survival::coxph(Surv(.) ~ variable, data, ...) |
| tbl_survfit() %>% add_p(test = X) | "coxph_wald" | Cox regression (Wald) | survival::coxph(Surv(.) ~ variable, data, ...) |
| tbl_survfit() %>% add_p(test = X) | "coxph_score" | Cox regression (Score) | survival::coxph(Surv(.) ~ variable, data, ...) |
| tbl_continuous() %>% add_p(test = X) | "anova_2way" | Two-way ANOVA | lm(continuous_variable ~ by + variable) |
| tbl_continuous() %>% add_p(test = X) | "t.test" | t-test | t.test(continuous_variable ~ as.factor(variable), data = data, conf.level = 0.95, ...) |
| tbl_continuous() %>% add_p(test = X) | "aov" | One-way ANOVA | aov(continuous_variable ~ as.factor(variable), data = data) %>% summary() |
| tbl_continuous() %>% add_p(test = X) | "kruskal.test" | Kruskal-Wallis test | kruskal.test(data[[continuous_variable]], as.factor(data[[variable]])) |
| tbl_continuous() %>% add_p(test = X) | "wilcox.test" | Wilcoxon rank-sum test | wilcox.test(as.numeric(continuous_variable) ~ as.factor(variable), data = data, ...) |
| tbl_continuous() %>% add_p(test = X) | "lme4" | random intercept logistic regression | lme4::glmer(by ~ (1 \UFF5C group), data, family = binomial) %>% anova(lme4::glmer(variable ~ continuous_variable + (1 \UFF5C group), data, family = binomial)) |
| tbl_continuous() %>% add_p(test = X) | "ancova" | ANCOVA | lm(continuous_variable ~ variable + adj.vars) |
| tbl_summary() %>% add_difference(test = X) | "t.test" | mean difference | t.test(variable ~ as.factor(by), data = data, conf.level = 0.95, ...) |
| tbl_summary() %>% add_difference(test = X) | "paired.t.test" | mean difference | tidyr::pivot_wider(id_cols = group, ...); t.test(by_1, by_2, paired = TRUE, conf.level = 0.95, ...) |
| tbl_summary() %>% add_difference(test = X) | "paired.wilcox.test" | rate difference | tidyr::pivot_wider(id_cols = group, ...); wilcox.test(by_1, by_2, paired = TRUE, conf.int = TRUE, conf.level = 0.95, ...) |
| tbl_summary() %>% add_difference(test = X) | "prop.test" | rate difference | prop.test(x, n, conf.level = 0.95, ...) |
| tbl_summary() %>% add_difference(test = X) | "ancova" | mean difference | lm(variable ~ by + adj.vars) |
| tbl_summary() %>% add_difference(test = X) | "ancova_lme4" | mean difference | lme4::lmer(variable ~ by + adj.vars + (1 \UFF5C group), data) |
| tbl_summary() %>% add_difference(test = X) | "cohens_d" | standardized mean difference | effectsize::cohens_d(variable ~ by, data, ci = conf.level, ...) |
| tbl_summary() %>% add_difference(test = X) | "smd" | standardized mean difference | smd::smd(x = data[[variable]], g = data[[by]], std.error = TRUE) |
| tbl_summary() %>% add_difference(test = X) | "emmeans" | adjusted mean difference | lm(variable ~ by + adj.vars, data) %>% emmeans::emmeans(specs =~by) %>% emmeans::contrast(method = "pairwise") %>% summary(infer = TRUE, level = conf.level) |
| tbl_svysummary() %>% add_difference(test = X) | "smd" | standardized mean difference | smd::smd(x = data$variables[[variable]], g = data$variables[[by]], w = weights(data), std.error = TRUE) |
少しでも、あなたの解析が楽になりますように!!